Adapted from this Brookhaven National Laboratory Press Release
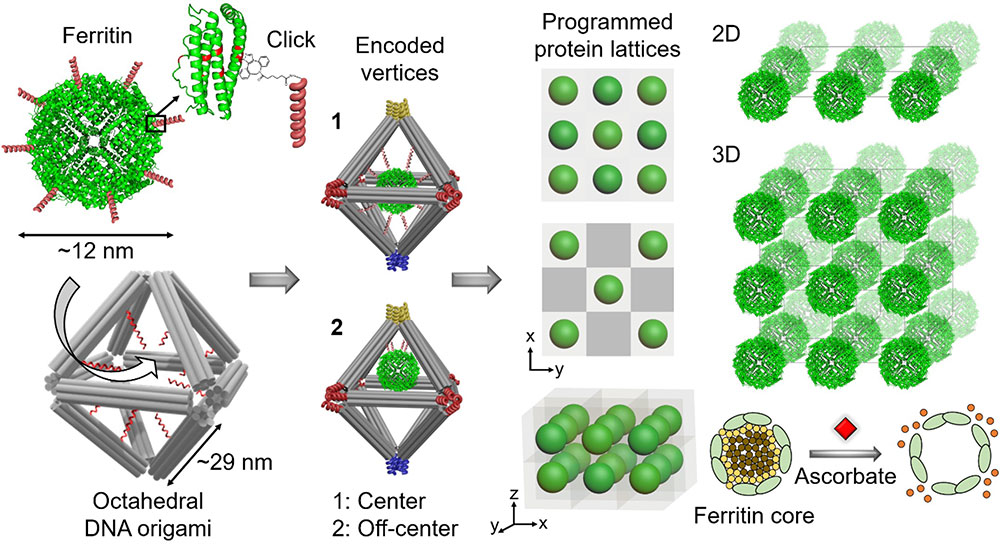
A team of Foundry users, working with staff, have organized proteins—nature’s most versatile building blocks—in desired 2-D and 3-D ordered arrays while maintaining their structural stability and biological activity. They built these designer functional protein arrays by using DNA as a programmable construction material. The team—representing the U.S. Department of Energy’s (DOE) Brookhaven National Laboratory, Columbia University, DOE’s Lawrence Berkeley National Laboratory, and City University of New York (CUNY)—described their approach in the June 17 issue of Nature Communications.
“For decades, scientists have dreamed about rationally assembling proteins into specific organizations with preserved protein function,” said corresponding author Oleg Gang, leader of the Center for Functional Nanomaterials (CFN) Soft and Bio Nanomaterials Group at Brookhaven Lab and a professor of chemical engineering and of applied physics and materials science at Columbia Engineering. “Our DNA-based platform has enormous potential not only for structural biology but also for various bioengineering, biomedical, and bionanomaterial applications.”
The primary motivation of this work was to establish a rational way to organize proteins into designed 2-D and 3-D architectures while preserving their function. The importance of organizing proteins is well known in the field of protein crystallography. For this technique, proteins are taken from their native solution-based environments and condensed to form an orderly arrangement of atoms (crystalline structure), which can then be structurally characterized. However, because of their flexibility and aggregation properties, many proteins are difficult to crystallize, requiring trial and error. The structure and function of proteins may change during the crystallization process, and they may become nonfunctional when crystallized by traditional methods. This new approach opens many possibilities for creating engineered biomaterials, beyond the goals of structural biology.
Though DNA is best known for its role in storing our genetic information, the very same base-pairing processes used for this storage can be leveraged to construct desired nanostructures. A single strand of DNA is made of subunits, or nucleotides, of which there are four kinds (known by the letters A, C, T, and G). Each nucleotide has a complementary nucleotide it attracts and binds to (A with T and C with G) when two DNA strands are near each other. Using this concept in the technique of DNA origami, scientists mix multiple short strands of synthetic DNA with a single long strand of DNA. The short strands bind to and “fold” the long strand into a particular shape based on the sequence of bases, which scientists can specify.
In this case, the scientists created octahedral-shaped DNA origami. Inside these cage-like frameworks, they placed DNA strands with a particular “color,” or coding sequence, at targeted locations (center and off center). To the surface of proteins—specifically, ferritin, which stores and releases iron, and apoferritin, its iron-free counterpart—they attached complementary DNA strands. By mixing the DNA cages and conjugated proteins and heating up the mixture to promote the reaction, the proteins went to the internal designated locations. They also created empty cages, without any protein inside.
To connect these nanoscale building blocks, or protein “voxels” (DNA cages with encapsulated proteins), in desired 2-D and 3-D arrays, second author and Columbia PhD student Brian Minevich designed different colors for the external bonds of the voxels. With this color scheme, the voxels would recognize each other in programmable, controllable ways leading to the formation of specifically prescribed types of protein lattices. To demonstrate the versatility of the platform, the team constructed single- and double-layered 2D arrays, as well as 3D arrays.
“By arranging the colors in a particular way, we can program the formation of different lattices,” explained Gang. “We have full control to design and build the protein lattice architectures we want.”enlarge
An illustration showing the approach for assembling biologically functional proteins into ordered 2-D and 3-D arrays through programmable octahedral-shaped DNA frameworks. These frameworks can host and control the placement of the proteins internally—for example, at the center (1) or off-center (2)—and be encoded with specific sequences externally (color coding scheme) to create desired 2-D and 3-D lattices. For example, red only connects to red, blue to blue, and so on. The team demonstrated the preserved biological activity of ferritin lattices by adding a compound (ascorbate) that induced the release of iron irons forming the ferritin core.
To confirm that the proteins had been encapsulated inside the cages and the lattices had been constructed as designed, the team turned to various electron- and x-ray-based imaging and scattering techniques. These techniques included electron microscopy (EM) imaging at the CFN; small-angle x-ray scattering at the National Synchrotron Light Source II (NSLS-II) Complex Materials Scattering (CMS) and Life Science X-ray Scattering (LiX) beamlines at Brookhaven; and cryogenic-EM imaging at the Molecular Foundry (MF) of Lawrence Berkeley and the CUNY Advanced Science Research Center. The CFN, NSLS-II, and MF are all DOE Office of Science User Facilities; CFN and MF are two of five DOE Nanoscale Science Research Centers.
“The science was enabled by advanced synthesis and characterization capabilities at three user facilities within the national lab system and one university-based facility,” said Gang. “Without these facilities and the expertise of scientists from each of them, this study wouldn’t have been possible.”enlarge
To visualize the assembled arrays in 3-D, the team applied nanoscale tomography based on the images obtained by cryo-electron microscopy. The top figure is a selected lattice area and the bottom a representative view. The color bars indicate different heights of the lattice (in angstroms).
Following these assembly studies, they investigated the biological activity of ferritin. By adding a reducing reagent to the ferritin lattice, they induced the release of iron ions from the center of the ferritin proteins.
“By monitoring the evolution of SAXS patterns during iron release, we could quantify how much iron was released and how quickly it was released, as well as confirm that the integrity of the lattice was maintained during this protein operation,” said Minevich. “According to our TEM studies, the proteins remained inside the frames.”
“We showed that the proteins can perform the same function as they do in a biological environment while keeping the spatial organization we created,” explained Wang.
Next, the team will apply their DNA-based platform to other types of proteins, with the goal of building more complex, operational protein systems.
“This research represents an important step in bringing together different components from real biological machinery and organizing them into desired 2-D and 3-D architectures to create engineered and bioactive materials,” said Gang. “It’s exciting because we see the rational path for fabricating desired functional bio-nano systems never-before produced by nature.”
This work was supported by the DOE Office of Science, a Laboratory Directed Research and Development Program grant, and the National Science Foundation. The LiX beamline is part of the Life Science Biomedical Technology Research resource, cofunded by the National Institute of General Medical Sciences and the DOE Office of Biological and Environmental Research, with additional support from the National Institutes of Health. Brookhaven’s Biology Department supplied the proteins.

