Peptoids are artificial versions of nature’s peptides that are highly customizable and have opened up new avenues of research in a range of fields, from biomedicine to materials science.
Peptoids can readily self-assemble into nanosheets, which are of particular interest because they are biocompatible, highly uniform bilayer structures that are free-floating in water. Nanosheets can serve as templates for the growth of composite materials and hold great potential for use as membranes for separations, or as a platform for chemical and biological sensing.
Recently, an international team of researchers discovered that peptoids can change shape when they form a nanosheet. Their findings were recently published in Proceedings of the National Academy of Sciences and The Journal of Physical Chemistry Letters.
The work was a collaboration between a group of Foundry researchers led by Ron Zuckermann and Steve Whitelam, and three teams of Foundry users from Georgia Tech, UC Irvine, and New York University Abu Dhabi.
“This project is a great example of the theory-experiment feedback cycle possible at the Foundry,” said Steve Whitelam, staff scientist in the Foundry’s Theory facility. “Experiments done by our users provided us with data that we used to improve our nanosheet models, which in turn allowed us to verify the consistency of our experimental data.”
In 2010, Ron Zuckermann and his group discovered a technique to synthesize peptoids into highly-ordered sheets that were just a few nanometers thick but up to 100 micrometers in length and width. Five years later, Whitelam led the development of the first computational molecular model of peptoid nanosheets that uncovered the design rule that makes peptoid nanosheets possible, and how their molecular architecture is closely related to a natural protein.
In this new work, Benjamin Hudson and Anant Paravastu, Foundry users from Georgia Tech, wanted to study the atomic details of the molecular structure of a peptoid nanosheet using the Foundry’s computational model in conjunction with a technique called solid-state nuclear magnetic resonance (NMR) spectroscopy.
At the Foundry, the researchers synthesized specially labelled peptoids that introduced “probe” atoms at specific locations within the molecule. Solid-state NMR showed exactly where the probes were in relation to each other, and revealed some surprising findings.
The Georgia Tech team found evidence supporting the brick-like pattern formed by peptoids in nanosheets predicted by the existing model, but also found that some of the bond angles inside individual peptoids did not match as expected.
With this new data in hand, the team used multi-scale simulations to create a new computational model. Whitelam and postdoctoral researcher John Edison worked with Ryan Spencer and Allon Hochbaum from UC Irvine, and Glenn Butterfoss from New York University Abu Dhabi, using the high-performance computing resources of Berkeley Lab’s National Energy Research Scientific Computing Center (NERSC).
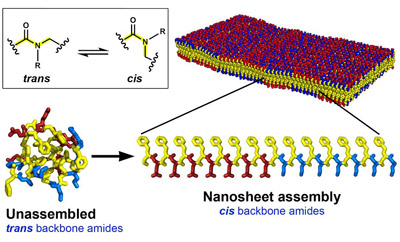
Version 2.0 of the Foundry’s model matched experimental data even better than before, and yielded a surprise. The reason that the bond angles didn’t match the original model is because peptoids can change shape when they assemble into nanosheets.
The shape of a molecule influences how it can be packed and arranged into a larger structure. When atoms are fixed in position on the same sides of a bond in a molecule, it is called “cis” (the name comes from latin, meaning “on this side”). If the atoms are fixed on opposite sides, then the structure is called “trans” (meaning “across”).
The backbone of individual peptoid molecules has a tendency to take on a trans configuration, because the structure is more energetically stable. However, the research team found that in nanosheets, the peptoids are primarily in the cis form.
When peptoids assemble into nanosheets, the “branches” of atoms coming off of the backbone interact with those of neighboring peptoids. These interactions become more favorable when the peptoid is in the cis position. While the peptoid backbone is less stable in the cis form, the atoms branching off of it become more stable and the molecules can be packed more tightly together and become more highly organized.
“In order to engineer nanosheets for biological or sensing applications we need to know the location of every peptoid atom in a nanosheet, “said John Edison, lead author of the article published in PNAS. “Prior to the Georgia Tech team’s work, we had no reason to believe that peptoids change shape when forming a nanosheet.”
With a better understanding of how peptoid chains organize themselves upon assembly, the researchers expect this work will open the door to creating new types of durable and engineerable biomimetic nanomaterials.

