by Clarissa Bhargava
Ask a biologist what the most complex molecules are in the body, and they’ll tell you: not DNA, but proteins. While your chromosomes are indeed the largest molecules in your body (comprised of billions of atoms), the structure of DNA itself is straightforward: two strands twisted into a double helix. Proteins, on the other hand, come in endless variety, their long chains folding in specific ways and carrying out every function of the body, from vision to digestion to DNA replication. At the Molecular Foundry, scientists have pioneered research into peptoids, a synthetic cousin of proteins, to engineer custom biomolecules with similarly breathtaking versatility, complexity, and biocompatibility.
In a study published last month, a research team led by Foundry alumna Jing Sun and Ron Zuckermann synthesized peptoid molecules with a unique hierarchical structure. A hallmark of proteins, predetermined higher-order structure is difficult to achieve in synthetic chemistry. The exact sequence of amino acids that forms a protein’s long backbone also determines how the molecule folds over itself to create precise nanostructures, which in turn determines how the protein functions. A single incorrect amino acid can throw the whole process off, but thanks to our DNA’s assembly instructions, most proteins come out perfect.
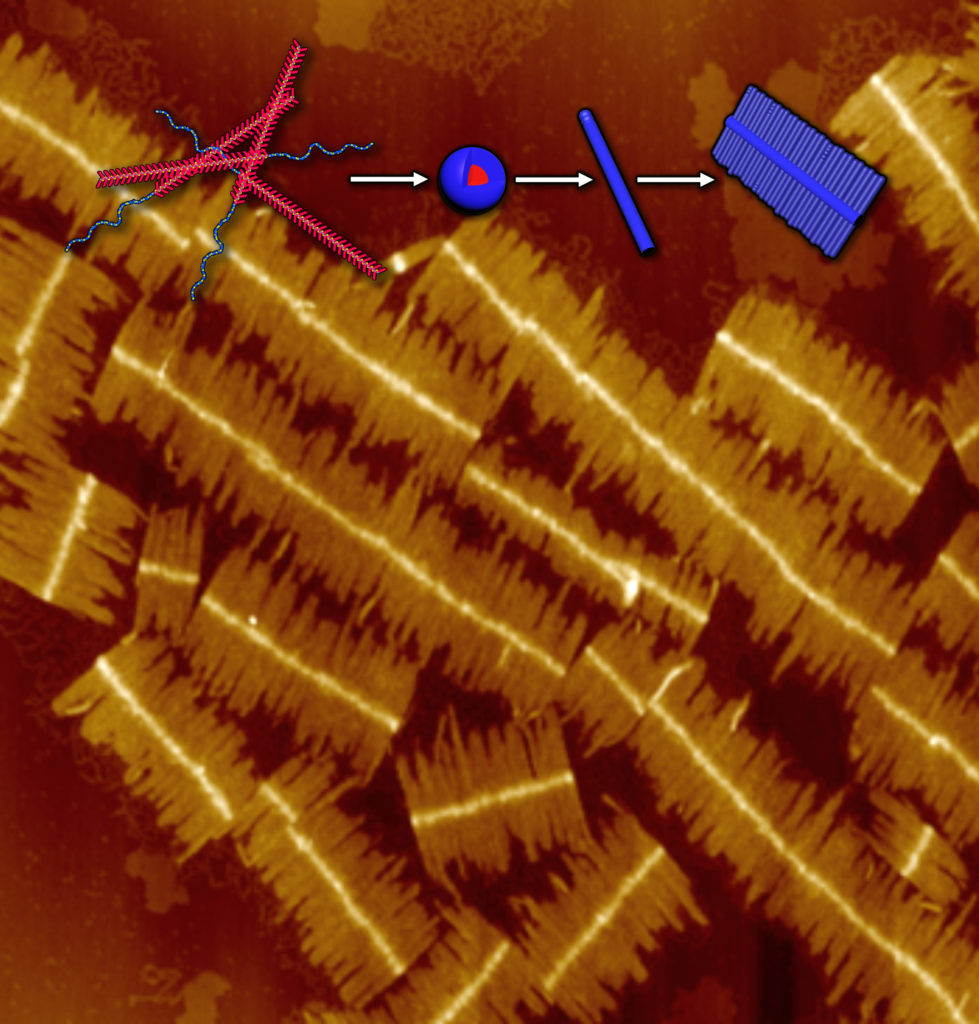
Making protein-like molecules from scratch is no easy task – it’s like building a palace without blueprints. Researchers have made use of advanced peptoid combinatorics, their synthesis savvy, and methodical experimental design in order to create brand-new peptoid architectures. Published in the journal PNAS, the recent study details peptoid “nanobrushes” composed of a single polymer species arranged in two distinct motifs, first forming a long “spine” and then assembling many shorter fibers on either side, like bristles on a brush.
The nanobrushes are made of a diblock copolymer – a long molecular chain with large, repeating stretches of each block – and are produced in three stages. First, the copolymer is assembled from its monomer subunits and dispersed in methanol. When the methanol suspension is warmed, tiny spheres called micelles form, and some of these spheres link up into long chains like a pearl necklace. These will become the spine of the nanobrush, reaching lengths of a micrometer or more. In the final synthesis step, the remaining spheres attach to the sides of the spine, growing outward to form thin lateral crystalline fibers in a process known as “living growth.”
By studying these nanobrushes with electron and atomic force microscopy, the research team was able to distinguish the detailed structures in both the spine and the lateral fibers. They also discovered that the brushes could be un-made and re-made many times by heating and cooling the solution again. At elevated temperatures, the lateral fibers pop off the spine intact, and at lower temperatures, they jump back on. Even more interesting, the final size and shape of the nanobrushes were found to be independent of the kinetic pathway taken, e.g., the cooling rate, making the nanobrush assembly a reversible, thermodynamically controlled process. This feature enables the researchers to adjust factors like the polymer concentration or the assembly time to control both the length and width of the brushes.
The successful design of the hierarchical nanobrush structures relied on scientists’ precise encoding of structural information into the polymer, essentially programming how the molecule will self-assemble into large-scale structures by making small-scale changes in the atomic sequence. By precisely balancing molecular controls like solubility and crystallinity, peptoid researchers can design biomimetic molecules for specific tasks from drug delivery to catalysis.
Once a postdoc at the Molecular Foundry, the study’s first author Jing Sun now leads her own peptoid research laboratory at Qingdao University of Science and Technology in China. The nanobrush research team was comprised of scientists from her lab, from the Chinese Academy of Sciences, and from Berkeley Lab’s ALS and Molecular Foundry. These collaborators are already working on their next paper, drafting custom blueprints for the emerging world of polypeptoid architecture.

Read the paper:
Jing Sun, Zhiwei Wang, Chenhui Zhu, Meiyao Wang, Zhekun Shi, Yuhan Wei, Xiaohui Fu, Xuesi Chen, Ronald N. Zuckermann. Proceedings of the National Academy of Sciences. Dec 2020, 117 (50) 31639-31647; DOI: 10.1073/pnas.2011816117

